● 앙상블(Ensamble)
- 일반화와 강건성(Robustness)을 향상시키기 위해 여러 모델의 예측 값을 결합하는 방법
- 앙상블의 종류
- 평균 방법
- 여러 추정값을 독립적으로 구한 뒤 평균을 취함
- 결합 추정값은 분산이 줄어들어 단일 추정값보다 좋은 성능
- 부스팅 방법
- 순차적으로 모델 생성
- 결합된 모델의 편향을 감소시키기 위해 노력
- 부스팅 방법의 목표는 여러 개의 약한 모델들을 결합해 하나의 강력한 앙상블 모델을 구축하는 것
- 평균 방법
1. Bagging meta-estimator
- bagging은 bootstrap arregatng의 줄임말
- 원래 훈련 데이터셋의 일부를 사용해 여러 모델을 훈련
- 각각의 결과를 결합해 최종 결과를 생성
- 분산을 줄이고 과적합을 막음
- 강력하고 복잡한 모델에서 잘 동작
- 사용 라이브러리 및 연습용 데이터 불러오기
from sklearn.datasets import load_iris, load_wine, load_breast_cancer, load_diabetes
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import cross_validate
from sklearn.ensemble import BaggingClassifier, BaggingRegressor
from sklearn.neighbors import KNeighborsClassifier, KNeighborsRegressor
from sklearn.svm import SVC, SVR
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
iris = load_iris()
wine = load_wine()
cancer = load_breast_cancer()
diabetes = load_diabetes()
- KNeighborsClassifier에서 일반 모델과 bagging 모델 비교
# 기본 모델 파이프라인 생성
base_model = make_pipeline(
StandardScaler(),
KNeighborsClassifier()
)
# bagging 모델 분류기 생성
bagging_model = BaggingClassifier(base_model, n_estimators = 10, max_samples = 0.5, max_features = 0.5)
# 일반 모델 파이프라인을 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = base_model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.0015646457672119141 (+/- 0.00046651220506644384)
avg score time: 0.0024621009826660155 (+/- 0.0007703127175682573)
avg test score: 0.9493650793650794 (+/- 0.037910929811115976)
# bagging 모델 분류기를 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = bagging_model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.01585836410522461 (+/- 0.001614824165009782)
avg score time: 0.004801464080810547 (+/- 0.0011524532393675253)
avg test score: 0.9496825396825397 (+/- 0.02695890511274157)- 일반 모델 파이프라인과 bagging 모델 분류기 간의 avg test score의 차이가 없음
- SVC에서 일반 모델과 bagging 모델 비교
# 기본 모델 파이프라인 생성
base_model = make_pipeline(
StandardScaler(),
SVC()
)
# bagging 모델 분류기 생성
bagging_model = BaggingClassifier(base_model, n_estimators = 10, max_samples = 0.5, max_features = 0.5)
# 일반 모델 파이프라인을 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = base_model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.0019928932189941405 (+/- 0.0006299217280194511)
avg score time: 0.0007997512817382813 (+/- 0.0003998954564704184)
avg test score: 0.9833333333333334 (+/- 0.022222222222222233)
# bagging 모델 분류기를 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = bagging_model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.03082098960876465 (+/- 0.010328177445939042)
avg score time: 0.003788471221923828 (+/- 0.0007431931234207308)
avg test score: 0.9495238095238095 (+/- 0.03680897280161461)- bagging 모델 분류기에서 avg test score가 일반 모델보다 더 감소함
- DecisionTreeClassifier에서 일반 모델과 bagging 모델 비교
# 기본 모델 파이프라인 생성
base_model = make_pipeline(
StandardScaler(),
DecisionTreeClassifier()
)
# bagging 모델 분류기 생성
bagging_model = BaggingClassifier(base_model, n_estimators = 10, max_samples = 0.5, max_features = 0.5)
# 일반 모델 파이프라인을 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = base_model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.0016411781311035157 (+/- 0.0005283725076633593)
avg score time: 0.0005584716796875 (+/- 0.00046257629871031806)
avg test score: 0.8709523809523809 (+/- 0.04130828490281938)
# bagging 모델 분류기를 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = bagging_model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.025203371047973634 (+/- 0.006259195949988035)
avg score time: 0.0019852161407470704 (+/- 0.0006092217277319406)
avg test score: 0.9665079365079364 (+/- 0.032368500562618134)- bagging 모델 분류기에서 avg test score가 일반 모델보다 더 증가함
- KNeighborsRegressor에서 일반 모델과 bagging 모델 비교
# 기본 모델 파이프라인 생성
base_model = make_pipeline(
StandardScaler(),
KNeighborsRegressor()
)
# bagging 모델 분류기 생성
bagging_model = BaggingClassifier(base_model, n_estimators = 10, max_samples = 0.5, max_features = 0.5)
# 일반 모델 파이프라인을 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = base_model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))# 출력 결과
# 출력 결과
avg fit time: 0.0011986732482910157 (+/- 0.0003976469573187139)
avg score time: 0.0011948585510253907 (+/- 0.0004032221568088893)
avg test score: 0.3689720650295623 (+/- 0.044659049060165365)
# bagging 모델 분류기를 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = bagging_model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))# 출력 결과
# 출력 결과
avg fit time: 0.01416783332824707 (+/- 0.0021502472754025685)
avg score time: 0.005353689193725586 (+/- 0.0005163540544834088)
avg test score: 0.4116223973880059 (+/- 0.039771045284647706)- bagging 모델 분류기에서 avg test score가 일반 모델보다 더 증가함
- SVR에서 일반 모델과 bagging 모델 비교
# 기본 모델 파이프라인 생성
base_model = make_pipeline(
StandardScaler(),
SVR()
)
# bagging 모델 분류기 생성
bagging_model = BaggingClassifier(base_model, n_estimators = 10, max_samples = 0.5, max_features = 0.5)
# 일반 모델 파이프라인을 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = base_model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.005189418792724609 (+/- 0.00039828050210851215)
avg score time: 0.0029881954193115234 (+/- 4.301525777064362e-05)
avg test score: 0.14659868748701582 (+/- 0.021908831719954277)
# bagging 모델 분류기를 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = bagging_model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))# 출력 결과
# 출력 결과
avg fit time: 0.030098438262939453 (+/- 0.004244103777765697)
avg score time: 0.014351558685302735 (+/- 0.002644062031057098)
avg test score: 0.06636804266664734 (+/- 0.026375606251683278)- bagging 모델 분류기에서 avg test score가 일반 모델보다 더 감소함
- DecisionTreeRegressor에서 일반 모델과 bagging 모델 비교
# 기본 모델 파이프라인 생성
base_model = make_pipeline(
StandardScaler(),
DecisionTreeRegressor()
)
# bagging 모델 분류기 생성
bagging_model = BaggingClassifier(base_model, n_estimators = 10, max_samples = 0.5, max_features = 0.5)
# 일반 모델 파이프라인을 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = base_model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.0030806541442871095 (+/- 0.0005084520543164849)
avg score time: 0.0005962371826171875 (+/- 0.0004868346584328474)
avg test score: -0.10402518188546787 (+/- 0.09614122612623877)
# bagging 모델 분류기를 사용하여 분류한 결과 정확도 출력
cross_val = cross_validate(
estimator = bagging_model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.028301239013671875 (+/- 0.009722365434744872)
avg score time: 0.001835775375366211 (+/- 0.0003816480339082081)
avg test score: 0.3206163477240517 (+/- 0.06240931175651393)- bagging 모델 분류기에서 avg test score가 일반 모델보다 더 증가함
- 데이터에 따라 다르지만 bagging 모델이 일반 모델보다 좋은 성능을 보임
2. Forests of randomized trees
- sklean.ensemble 모듈에는 무작위 결정 트리를 기반으로 하는 두 개의 평균화 알고리즘이 존재
- Random Forest
- Extra-Trees
- 모델 구성에 임의성을 추가해 다양한 모델 집합 생성
- 앙상블 모델의 예측은 각 모델의 평균
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier
- Random Forests 분류
# 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
RandomForestClassifier()
)
# 붓꽃 데이터
cross_val = cross_validate(
estimator = model,
X = iris.data, y = iris.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.18105216026306153 (+/- 0.01685682915874488)
avg score time: 0.013797760009765625 (+/- 0.0025621248659456705)
avg test score: 0.96 (+/- 0.024944382578492935)
# 와인 데이터
cross_val = cross_validate(
estimator = model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.22976922988891602 (+/- 0.028256732778271565)
avg score time: 0.01587204933166504 (+/- 0.0023274044664123553)
avg test score: 0.9665079365079364 (+/- 0.032368500562618134)
# 유방암 데이터
cross_val = cross_validate(
estimator = model,
X = cancer.data, y = cancer.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.2917177200317383 (+/- 0.045315794078103835)
avg score time: 0.017597723007202148 (+/- 0.006803860063290489)
avg test score: 0.9578326346840551 (+/- 0.02028739541529243)
- Random Forests 회귀
# 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
RandomForestRegressor()
)
# 당뇨병 데이터
cross_val = cross_validate(
estimator = model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.38582072257995603 (+/- 0.05492288277511406)
avg score time: 0.01399979591369629 (+/- 0.0017906758222056265)
avg test score: 0.41577582207684943 (+/- 0.04029500816412448)
- Extremely Randomized Trees 분류
# 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
ExtraTreesClassifier()
)
# 붓꽃 데이터
cross_val = cross_validate(
estimator = model,
X = iris.data, y = iris.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.12993454933166504 (+/- 0.013365132836320116)
avg score time: 0.015802621841430664 (+/- 0.0026369354952103644)
avg test score: 0.9533333333333334 (+/- 0.03399346342395189)
# 와인 데이터
cross_val = cross_validate(
estimator = model,
X = cancer.data, y = cancer.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.14599318504333497 (+/- 0.027877641431797804)
avg score time: 0.013452291488647461 (+/- 0.0019263323264865461)
avg test score: 0.9776190476190475 (+/- 0.020831783767013237)
# 유방암 데이터
cross_val = cross_validate(
estimator = model,
X = cancer.data, y = cancer.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.15427541732788086 (+/- 0.01803722986834076)
avg score time: 0.014791202545166016 (+/- 0.0007367600639162756)
avg test score: 0.9683744760130415 (+/- 0.010503414750935476)
- Extremely Randomized Trees 회귀
# 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
ExtraTreesRegressor()
)
# 당뇨병 데이터
cross_val = cross_validate(
estimator = model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.21371040344238282 (+/- 0.029887789885253244)
avg score time: 0.011214303970336913 (+/- 0.0022113466437151)
avg test score: 0.4334859913753323 (+/- 0.037205109491594564)
- Random Forest, Extra Tree 시각화
- 결정 트리, Random Forest, Extra Tree의 결정 경계와 회귀식 시각화
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.tree import DecisionTreeClassifier
# 기본 파라미터
n_classes = 3
n_estimators = 30
cmap = plt.cm.RdYlBu
plot_step = 0.02
plot_step_coarser = 0.5
RANDOM_SEED = 13
iris = load_iris()
plot_idx = 1
# 세 가지 모델을 for문에 넣어서 각 모델에 대한 예측 결과를 출력
models = [DecisionTreeClassifier(max_depth = None),
RandomForestClassifier(n_estimators = n_estimators),
ExtraTreesClassifier(n_estimators = n_estimators)]
plt.figure(figsize = (16, 8))
for pair in ([0, 1], [0, 2], [2, 3]):
# 위에서 지정한 세가지 모델에 대해 각각 예측
for model in models:
X = iris.data[:, pair]
y = iris.target
# 독립변수의 개수만큼 인덱스 배열 생성
idx = np.arange(X.shape[0])
# 랜덤시드 설정하고 랜덤으로 인덱스 셔플링(필요 x)
np.random.seed(RANDOM_SEED)
np.random.shuffle(idx)
X = X[idx]
y = y[idx]
# 평균과 표준편차를 계산하여 데이터 정규화
mean = X.mean(axis = 0)
std = X.std(axis = 0)
X = (X-mean) / std
# 위에서 정규화한 X와 y값을 모델에 피팅
model.fit(X, y)
# 모델 제목은 모델의 타입을 "."으로 분리하여
# ["<class 'sklearn", 'tree', '_classes', "DecisionTreeClassifier'>"]
# [-1]은 리스트의 가장 마지막 부분 추출("DecisionTreeClassifier'>")
# [:-2]는 위의 문자열의 끝에서 두번째 문자전까지만 추출("DecisionTreeClassifier")
# 마지막으로 해당 문자열에서 Classifier을 제외하기 위해 Classifier의 문자열 길이 전까지만 추출
model_title = str(type(model)).split(".")[-1][:-2][:-len("Classifier")]
# 3 * 3의 그래프 배열에서 plot_idx만큼(plot_idx는 모델 한 번 돌때마다 변경됨)
plt.subplot(3, 3, plot_idx)
if plot_idx <= len(models):
plt.title(model_title, fontsize = 9)
# meshgrid 생성
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, plot_step),
np.meshgrid(np.arange(y_min, y_max, plot_step)))
# DecisionTreeClassifier 모델일 때와 나머지 모델일 때 구분
if isinstance(model, DecisionTreeClassifier):
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cs = plt.contourf(xx, yy, Z, cmap = cmap)
# 나머지 모델일 때는 alpha값을 주어 투명도 추가
else:
estimator_alpha = 1.0 / len(model.estimators_)
for tree in model.estimators_:
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cs = plt.contourf(xx, yy, Z, alpha = estimator_alpha, cmap = cmap)
xx_coarser, yy_coarser = np.meshgrid(np.arange(x_min, x_max, plot_step_coarser),
np.arange(y_min, y_max, plot_step_coarser))
Z_points_coarser = model.predict(np.c_[xx_coarser.ravel(),
yy_coarser.ravel()]).reshape(xx_coarser.shape)
cs_points = plt.scatter(xx_coarser, yy_coarser, s = 15, c = Z_points_coarser, cmap = cmap, edgecolor = 'none')
plt.scatter(X[:, 0], X[:, 1], c = y, cmap = ListedColormap(['r','y','b']), edgecolor = 'k', s = 20)
plot_idx += 1
# 그래프 최상단 제목 설정
plt.suptitle("Classifiers", fontsize = 12)
plt.axis('tight')
plt.tight_layout(h_pad = 0.2, w_pad = 0.2, pad = 2.5)
plt.show()
plot_idx = 1
models = [DecisionTreeRegressor(max_depth = None),
RandomForestRegressor(n_estimators = n_estimators),
ExtraTreesRegressor(n_estimators = n_estimators)]
plt.figure(figsize = (16, 8))
for pair in (0, 1, 2):
for model in models:
X = diabetes.data[:, pair]
y = diabetes.target
idx = np.arange(X.shape[0])
np.random.seed(RANDOM_SEED)
np.random.shuffle(idx)
X = X[idx]
y = y[idx]
mean = X.mean(axis = 0)
std = X.std(axis = 0)
X = (X-mean) / std
model.fit(X.reshape(-1, 1), y)
model_title = str(type(model)).split(".")[-1][:-2][:-len('Regreffor')]
plt.subplot(3, 3, plot_idx)
if plot_idx <= len(models):
plt.title(model_title, fontsize = 9)
x_min, x_max = X.min() -1, X.max() + 1
y_min, y_max = y.min() -1, y.max() + 1
xx, yy = np.arange(x_min - 1, x_max + 1, plot_step), np.arange(y_min - 1, y_max + 1, plot_step)
if isinstance(model, DecisionTreeRegressor):
Z = model.predict(xx.reshape(-1, 1))
cs = plt.plot(xx, Z)
else:
estimator_alpha = 1.0 / len(model.estimators_)
for tree in model.estimators_:
Z = tree.predict(xx.reshape(-1, 1))
cs = plt.plot(xx, Z, alpha = estimator_alpha)
plt.scatter(X, y, edgecolors = 'k', s = 20)
plot_idx += 1
plt.suptitle("Regressor", fontsize = 12)
plt.axis('tight')
plt.tight_layout(h_pad = 0.2, w_pad = 0.2, pad = 2.5)
plt.show()
3. AdaBoost
- 대표적인 부스팅 알고리즘
- 일련의 약한 모델들을 학습
- 수정된 버전의 데이터를 (가중치가 적용된)반복 학습
- 가중치 투표(또는 합)을 통해 각 모델의 예측값을 결합
- 첫 단계에서는 원본 데이터를 학습하고 연속적인 반복마다 개별 샘플에 대한 가중치가 수정되고 다시 모델이 학습
- 잘못 예측된 샘플은 가중치가 증가, 올바르게 예측된 샘플은 가중치 감소
- 각각의 약한 모델들은 예측하기 어려운 샘플에 집중하게 됨
- AdaBoostClassifier
from sklearn.ensemble import AdaBoostClassifier, AdaBoostRegressor
# 기본 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
AdaBoostClassifier()
)
# 붓꽃 데이터
cross_val = cross_validate(
estimator = model,
X = iris.data, y = iris.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.10685276985168457 (+/- 0.0251679732383264)
avg score time: 0.012082815170288086 (+/- 0.0029404438972186714)
avg test score: 0.9466666666666667 (+/- 0.03399346342395189)
# 와인 데이터
cross_val = cross_validate(
estimator = model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.12471566200256348 (+/- 0.022184160168356563)
avg score time: 0.010206842422485351 (+/- 0.0014569535430837709)
avg test score: 0.8085714285714285 (+/- 0.16822356718459935)
# 유방암 데이터
cross_val = cross_validate(
estimator = model,
X = cancer.data, y = cancer.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.21773457527160645 (+/- 0.036282505551639664)
avg score time: 0.01280508041381836 (+/- 0.006666702833190784)
avg test score: 0.9701133364384411 (+/- 0.019709915473893072)
- AdaBoostRegressor
from sklearn.ensemble import AdaBoostClassifier, AdaBoostRegressor
# 기본 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
AdaBoostRegressor()
)
# 당뇨병 데이터
cross_val = cross_validate(
estimator = model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.0806793212890625 (+/- 0.019655713051304806)
avg score time: 0.004206609725952148 (+/- 0.0017171964676314028)
avg test score: 0.4325980577698525 (+/- 0.046753912177138576)
4. Gradient Tree Boosting
- 임의의 차별화 가능한 손실함수로 일반화한 부스팅 알고리즘
- 웹 검색, 분류 및 회귀 등 다양한 분야에서 모두 사용 가능
- GradientBoostingClassifier
from sklearn.ensemble import GradientBoostingClassifier, GradientBoostingRegressor
# 기본 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
GradientBoostingClassifier()
)
# 붓꽃 데이터
cross_val = cross_validate(
estimator = model,
X = iris.data, y = iris.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.28792128562927244 (+/- 0.08891755551147741)
avg score time: 0.0009984493255615235 (+/- 1.4898700510940572e-05)
avg test score: 0.9666666666666668 (+/- 0.02108185106778919)
# 와인 데이터
cross_val = cross_validate(
estimator = model,
X = wine.data, y = wine.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.557232141494751 (+/- 0.11776815518280624)
avg score time: 0.001400327682495117 (+/- 0.0004905852751874076)
avg test score: 0.9385714285714286 (+/- 0.032068206474093704)
# 유방암 데이터
avg fit time: 0.5082685947418213 (+/- 0.05271622631546759)
avg score time: 0.0009903907775878906 (+/- 9.857966772333804e-06)
avg test score: 0.9596180717279925 (+/- 0.02453263202329889)
- GradientBoostingRegressor
from sklearn.ensemble import GradientBoostingClassifier, GradientBoostingRegressor
# 기본 모델 파이프라인 생성
model = make_pipeline(
StandardScaler(),
GradientBoostingRegressor()
)
# 당뇨병 데이터
cross_val = cross_validate(
estimator = model,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print("avg fit time: {} (+/- {})".format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print("avg score time: {} (+/- {})".format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print("avg test score: {} (+/- {})".format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.12577242851257325 (+/- 0.01002747346482568)
avg score time: 0.0013751983642578125 (+/- 0.00048391264033975266)
avg test score: 0.40743256738384626 (+/- 0.06957393690515927)
5. 투표 기반 분류(Voting Classifier)
- 서로 다른 모델들의 결과를 투표를 통해 결합
- 두가지 방법으로 투표 가능
- 가장 많이 예측된 클래스를 정답으로 채택(hard voting)
- 예측된 확률의 가중치 평균(soft voting)
- hard 보팅
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import VotingClassifier
from sklearn.model_selection import cross_val_score
model1 = SVC()
model2 = GaussianNB()
model3 = RandomForestClassifier()
vote_model = VotingClassifier(
estimators = [('svc', model1), ('naive', model2), ('forest', model3)],
voting = 'hard'
)
for model in (model1, model2, model3, vote_model):
model_name = str(type(model)).split(".")[-1][:-2]
scores = cross_val_score(model, iris.data, iris.target, cv = 5)
print('accurancy: %0.2f (+/- %0.2f) [%s]' % (scores.mean(), scores.std(), model_name))
# 출력 결과
accurancy: 0.97 (+/- 0.02) [SVC]
accurancy: 0.95 (+/- 0.03) [GaussianNB]
accurancy: 0.96 (+/- 0.02) [RandomForestClassifier]
accurancy: 0.96 (+/- 0.02) [VotingClassifier]
- soft 보팅
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import VotingClassifier
from sklearn.model_selection import cross_val_score
model1 = SVC(probability = True)
model2 = GaussianNB()
model3 = RandomForestClassifier()
vote_model = VotingClassifier(
estimators = [('svc', model1), ('naive', model2), ('forest', model3)],
voting = 'soft',
weights = [2, 1, 2]
)
for model in (model1, model2, model3, vote_model):
model_name = str(type(model)).split(".")[-1][:-2]
scores = cross_val_score(model, iris.data, iris.target, cv = 5)
print('accurancy: %0.2f (+/- %0.2f) [%s]' % (scores.mean(), scores.std(), model_name))
# 출력 결과
accurancy: 0.97 (+/- 0.02) [SVC]
accurancy: 0.95 (+/- 0.03) [GaussianNB]
accurancy: 0.97 (+/- 0.02) [RandomForestClassifier]
accurancy: 0.96 (+/- 0.02) [VotingClassifier]
- 결정경계 시각화
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import VotingClassifier
from itertools import product
# 총 4개의 feature 중 2개만 사용
X = iris.data[:, [0, 2]]
y = iris.target
model1 = DecisionTreeClassifier(max_depth = 4)
model2 = KNeighborsClassifier(n_neighbors = 7)
model3 = SVC(gamma = .1, kernel = 'rbf', probability = True)
vote_model = VotingClassifier(estimators = [('dt', model1), ('knn', model2), ('svc', model3)], voting = 'soft', weights = [2, 1, 2])
model1 = model1.fit(X, y)
model2 = model2.fit(X, y)
model3 = model3.fit(X, y)
vote_model = vote_model.fit(X, y)
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.1),
np.arange(y_min, y_max, 0.1))
f, axarr = plt.subplots(2, 2, sharex = 'col', sharey = 'row', figsize = (12, 8))
for idx, model, tt in zip(product([0, 1], [0,1]), [model1, model2, model3, vote_model],
['Decision Tree (depth=4)', 'KNN (k=7)', 'Kernel SVM', 'Soft Voting']):
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
axarr[idx[0], idx[1]].contourf(xx, yy, Z, alpha = 0.4)
axarr[idx[0], idx[1]].scatter(X[:, 0], X[:, 1], c = y, s = 20, edgecolor = 'k')
axarr[idx[0], idx[1]].set_title(tt)
plt.show()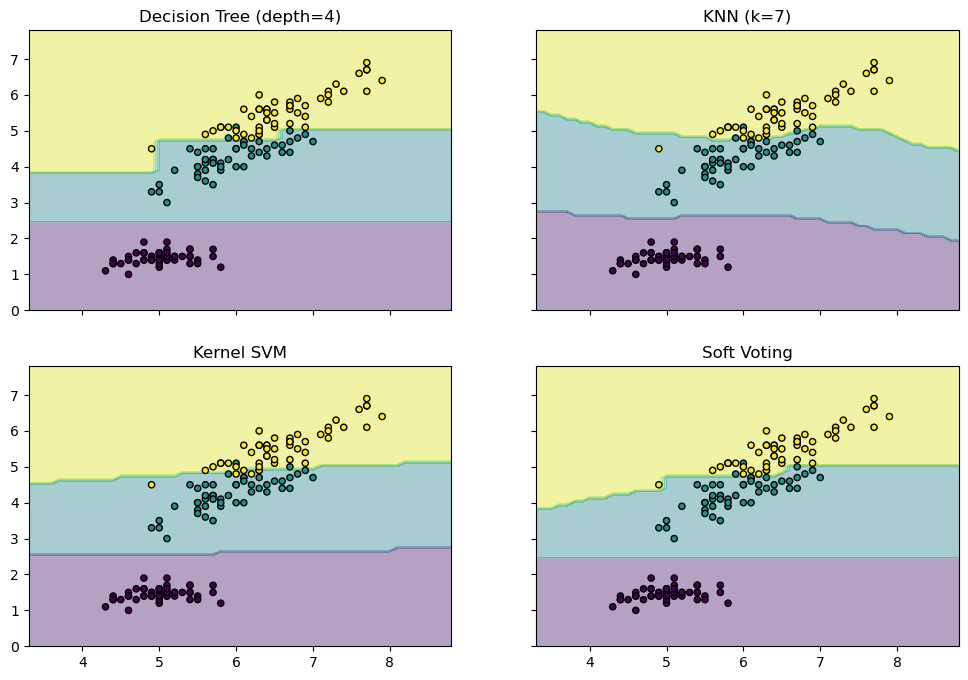
6. 투표 기반 회귀
- 서로 다른 모델의 예측값의 평균을 사용
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import GradientBoostingRegressor, RandomForestRegressor, VotingRegressor
model1 = LinearRegression()
model2 = GradientBoostingRegressor()
model3 = RandomForestRegressor()
vote_model = VotingRegressor(
estimators = [('Linear', model1), ('gbr', model2), ('rfr', model3)],
weights = [1, 1, 1]
)
for model in (model1, model2, model3, vote_model):
model_name = str(type(model)).split(".")[-1][:-2]
scores = cross_val_score(model, diabetes.data, diabetes.target, cv = 5)
print('R2: %0.2f (+/- %0.2f) [%s]' % (scores.mean(), scores.std(), model_name))
# 출력 결과
R2: 0.48 (+/- 0.05) [LinearRegression]
R2: 0.41 (+/- 0.07) [GradientBoostingRegressor]
R2: 0.42 (+/- 0.05) [RandomForestRegressor]
R2: 0.46 (+/- 0.05) [VotingRegressor]
- 시각화
X = diabetes.data[:, 0].reshape(-1, 1)
y = diabetes.target
model1 = LinearRegression()
model2 = GradientBoostingRegressor()
model3 = RandomForestRegressor()
vote_model = VotingRegressor(
estimators = [('Linear', model1), ('gbr', model2), ('rfr', model3)],
weights = [1, 1, 1]
)
model1 = model1.fit(X, y)
model2 = model2.fit(X, y)
model3 = model3.fit(X, y)
vote_model = vote_model.fit(X, y)
x_min, x_max = X.min() - 1, X.max() + 1
xx = np.arange(x_min - 1, x_max + 1, 0.1)
f, axarr = plt.subplots(2, 2, sharex = 'col', sharey = 'row', figsize = (12, 8))
for idx, model, tt in zip(product([0, 1], [0, 1]),
[model1, model2, model3, vote_model],
['Linear Regressor', 'Gradient Boosting', 'Random Forest', 'Voting']):
Z = model.predict(xx.reshape(-1, 1))
axarr[idx[0], idx[1]].plot(xx, Z, c = 'r')
axarr[idx[0], idx[1]].scatter(X, y, s = 20, edgecolor = 'k')
axarr[idx[0], idx[1]].set_title(tt)
plt.show()
7. 스택 일반화(Stacked Generalization)
- 각 모델의 예측값을 최종 모델의 입력으로 사용(모델의 예측값을 최종 모델에 스택으로 쌓음)
- 모델의 편향을 줄이는데 효과적
- 스택 회귀
from sklearn.linear_model import Ridge, Lasso
from sklearn.svm import SVR
from sklearn.ensemble import GradientBoostingRegressor, StackingRegressor
estimators = [('ridge', Ridge()),
('lasso', Lasso()),
('svr', SVR())]
reg = make_pipeline(
StandardScaler(),
StackingRegressor(estimators = estimators,
final_estimator = GradientBoostingRegressor())
)
cross_val = cross_validate(
estimator = reg,
X = diabetes.data, y = diabetes.target,
cv = 5
)
print('avg fit time: {} (+/- {})'.format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print('avg score time: {} (+/- {})'.format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print('avg test score: {} (+/- {})'.format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 0.34942941665649413 (+/- 0.10878244133017631)
avg score time: 0.010065841674804687 (+/- 0.00266810792836556)
avg test score: 0.37135451119632856 (+/- 0.0688832914154432)
- 회귀식 시각화
X = diabetes.data[:, 0].reshape(-1, 1)
y = diabetes.target
model1 = Ridge()
model2 = Lasso()
model3 = SVR()
reg = StackingRegressor(estimators = estimators,
final_estimator = GradientBoostingRegressor())
model1 = model1.fit(X, y)
model2 = model2.fit(X, y)
model3 = model3.fit(X, y)
reg = reg.fit(X, y)
x_min, x_max = X.min() - 1, X.max() + 1
xx = np.arange(x_min - 1, x_max + 1, 0.1)
f, axarr = plt.subplots(2, 2, sharex = 'col', sharey = 'row', figsize = (12, 8))
for idx, model, tt in zip(product([0, 1], [0, 1]),
[model1, model2, model3, vote_model],
['Ridge', 'Lasso', 'SVR', 'Stack']):
Z = model.predict(xx.reshape(-1, 1))
axarr[idx[0], idx[1]].plot(xx, Z, c = 'r')
axarr[idx[0], idx[1]].scatter(X, y, s = 20, edgecolor = 'k')
axarr[idx[0], idx[1]].set_title(tt)
plt.show()
- 스택 분류
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import StackingClassifier
estimators = [('logistic', LogisticRegression(max_iter = 10000)),
('svc', SVC()),
('naive', GaussianNB())]
clf = StackingClassifier(
estimators = estimators,
final_estimator = RandomForestClassifier()
)
cross_val = cross_validate(
estimator = clf,
X = iris.data, y = iris.target,
cv = 5
)
print('avg fit time: {} (+/- {})'.format(cross_val['fit_time'].mean(), cross_val['fit_time'].std()))
print('avg score time: {} (+/- {})'.format(cross_val['score_time'].mean(), cross_val['score_time'].std()))
print('avg test score: {} (+/- {})'.format(cross_val['test_score'].mean(), cross_val['test_score'].std()))
# 출력 결과
avg fit time: 1.7309207439422607 (+/- 0.5153520045243725)
avg score time: 0.08048839569091797 (+/- 0.06365540285519627)
avg test score: 0.9733333333333334 (+/- 0.02494438257849294)
- 결정 경계 시각화
# 총 4개의 feature 중 2개만 사용
X = iris.data[:, [0, 2]]
y = iris.target
model1 = LogisticRegression(max_iter = 10000)
model2 = SVC()
model3 = GaussianNB()
stack = StackingClassifier(estimators = estimators, final_estimator = RandomForestClassifier())
model1 = model1.fit(X, y)
model2 = model2.fit(X, y)
model3 = model3.fit(X, y)
stack = stack.fit(X, y)
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.1),
np.arange(y_min, y_max, 0.1))
f, axarr = plt.subplots(2, 2, sharex = 'col', sharey = 'row', figsize = (12, 8))
for idx, model, tt in zip(product([0, 1], [0,1]), [model1, model2, model3, stack],
['Logistic Regression', 'SVC', 'GaussianNB', 'Stack']):
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
axarr[idx[0], idx[1]].contourf(xx, yy, Z, alpha = 0.4)
axarr[idx[0], idx[1]].scatter(X[:, 0], X[:, 1], c = y, s = 20, edgecolor = 'k')
axarr[idx[0], idx[1]].set_title(tt)
plt.show()
'Python > Machine Learning' 카테고리의 다른 글
| [머신러닝 알고리즘] 군집화 (1) | 2023.03.02 |
|---|---|
| [머신러닝 알고리즘] XGBoost, LightGBM (0) | 2023.02.23 |
| [머신러닝 알고리즘] 결정 트리 (0) | 2023.01.18 |
| [머신러닝 알고리즘] 서포트 벡터 머신 (0) | 2023.01.13 |
| [머신러닝 알고리즘] 나이브 베이즈 (0) | 2023.01.10 |